Simultaneous multi-color 3D whole cell super-resolution imaging and particle tracking using engineered point spread functions
- Abstract number
- 32
- Presentation Form
- Poster
- DOI
- 10.22443/rms.elmi2021.32
- Corresponding Email
- [email protected]
- Session
- Poster Session 2
- Authors
- Anurag Agrawal (1), Warren Colomb (1), Scott Gaumer (1), Anjul Loiacono (1), Leslie Kimerling (1)
- Affiliations
-
1. Double Helix Optics
- Keywords
Multi-color 3D imaging; Point Spread Function Engineering; Double Helix PSF; 3D Particle Tracking; 3D Superresolution
- Abstract text
Widefield fluorescence optical microscopes are the workhorse of laboratories in numerous fields – biology, medicine, and pharmacology – providing invaluable visual information and informative quantitative data. The function of bio-molecules is often inferred by comparing the behavior of different intercellular functions by labeling them with different molecules. It is therefore valuable to localize and track multiple colors simultaneously in 3D. However, currently available microscopes are often limited in their imaging depth, resolution, and ability to image different fluorophores simultaneously.
Current solutions rely on sequential imaging or an additional camera and do not provide the required flexibility to work with the recently available large sensor formats (25 mm diagonal) and the ubiquitous legacy format (11.6 mm diagonal). Furthermore, none of the existing solutions take advantage of the increased depth, speed, and 3D resolution enhancements provided by engineered PSFs[1]–[3]. Here, we present two unique solutions that overcome these limitations and enable simultaneous 3D extended depth multi-color imaging of biological samples.
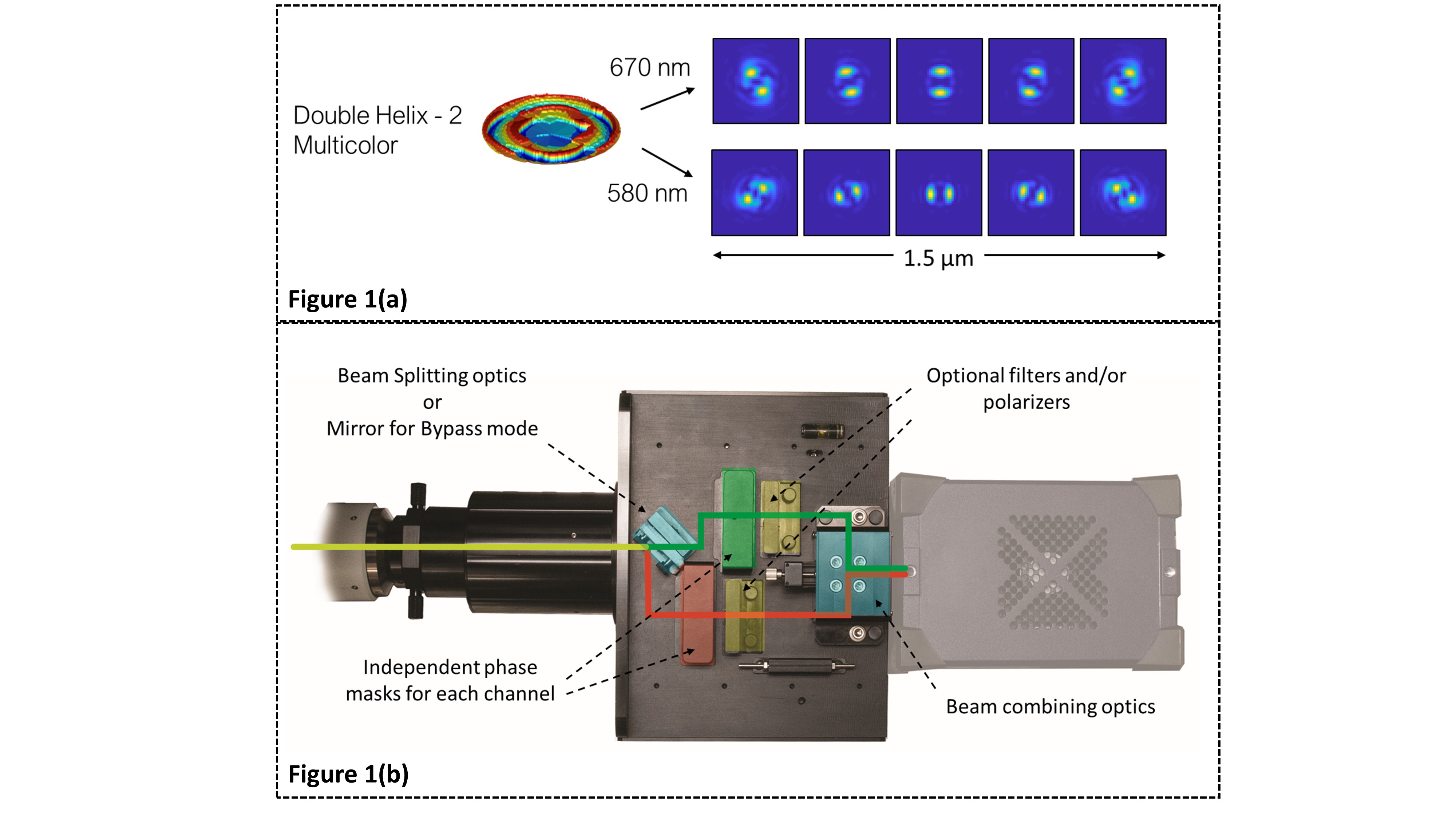
We address the limitations of existing solutions in two ways. Our first solution is based on multi-color phase masks (MC-PM) [4], which uniquely identify two or more probes on the same camera sensor area by encoding the spectral information and axial position, into a distinctly-shaped PSF for each color. An example of this is shown in Fig 1 where the MC-PM encodes DH-PSFs at perpendicular orientations for two selected wavelengths.
Our second solution is a universal modular subsystem – SPINDLE2, which can be installed between most scientific microscopes and cameras (see Figure 2). Along with a library of engineered PSFs, including MC-PMs[4], Double-Helix[1], and Tetrapod[3], the SPINDLE2 can be used with dichroic mirrors or polarizing/non-polarizing beam splitters. It is easily adjusted to work with a wide range of camera formats with no loss of resolution across the full field of view. The module features interchangeable mounts enabling users to optimize the PSF for their experiments, while allowing easy switching between single color, dual color, and bypass modes. The library of phase masks allows users to choose from a broad range of PSFs to extend the imaging depth range by 3x-30x, allowing for up to 30 µm of depth range with high numerical aperture objectives.
The unique combination of multimodal imaging and engineered PSFs has been used to extend the imaging capabilities of microscopes in the areas of nanometer-scale single-molecule imaging[1], [2], light sheet[5], 3D particle tracking [3], [4], the study of cancer[6], immunology, neuroscience[7], and more.
Figure 1: (a) A multicolor Double Helix mask exhibiting unique point spread functions based on the emission wavelength of the fluorophore enabling simultaneous multicolor imaging (b) Schematic of the Double Helix SPINDLE2 module showing the beam paths of each channel. The module allows for simultaneous imaging of two channels (emission spectrum based, polarization based, intensity based, etc.) on a single camera. Each imaging channel can be modulated with a point spread function from the Double Helix library of phase masks.
- References
[1] S. R. P. Pavani et al., “Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function.,” Proc. Natl. Acad. Sci. U. S. A., vol. 106, no. 9, pp. 2995–9, Mar. 2009.
[2] A. Gahlmann et al., “Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions.,” Nano Lett., vol. 13, no. 3, pp. 987–93, Mar. 2013.
[3] Y. Shechtman, L. E. Weiss, A. S. Backer, S. J. Sahl, and W. E. Moerner, “Precise Three-Dimensional Scan-Free Multiple-Particle Tracking over Large Axial Ranges with Tetrapod Point Spread Functions,” Nano Lett., vol. 15, no. 6, pp. 4194–4199, Jun. 2015.
[4] Y. Shechtman, L. E. Weiss, A. S. Backer, M. Y. Lee, and W. E. Moerner, “Multicolour localization microscopy by point-spread-function engineering,” Nat. Photonics, vol. 10, no. 9, pp. 590–594, Sep. 2016.
[5] A.-K. Gustavsson, P. N. Petrov, M. Y. Lee, Y. Shechtman, and W. E. Moerner, “3D single-molecule super-resolution microscopy with a tilted light sheet,” Nat. Commun. 2018 91, vol. 9, no. 1, p. 123, Jan. 2018.
[6] W. Y. Lam et al., “HER2 Cancer Protrusion Growth Signaling Regulated by Unhindered, Localized Filopodial Dynamics,” bioRxiv, p. 654988, Jun. 2019.
[7] S. Jain, J. R. Wheeler, R. W. Walters, A. Agrawal, A. Barsic, and R. Parker, “ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure,” Cell, vol. 164, no. 3, pp. 487–498, 2016.