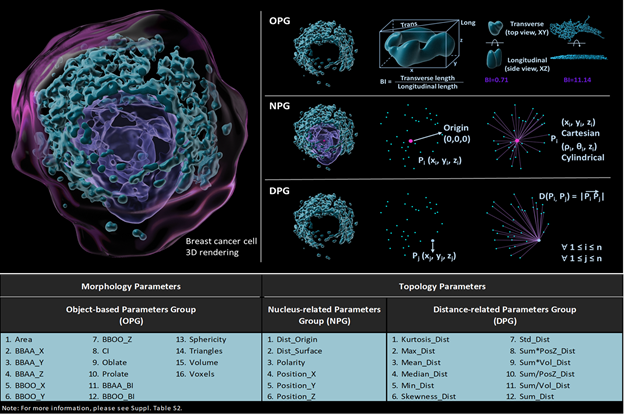
Organelle topology is a new breast cancer cell classifier
- Abstract number
- 72
- Presentation Form
- Poster Flash Talk and Poster
- DOI
- 10.22443/rms.elmi2021.72
- Corresponding Email
- [email protected]
- Session
- Poster Session 2
- Authors
- Ling Wang (1), Joshua Goldwag (1), Megan Bouyea (1), Jamie Ward (1), Niva Maharjan (2), Amina Eladdadi (2), Margarida Barroso (1)
- Affiliations
-
1. Albany Medical College
2. The College of Saint Rose
- Keywords
organelles, morphology, topology, machine learning, endosomes, mitochondria
- Abstract text
Breast cancer is a highly heterogeneous disease, both phenotypically and genetically. The spatial organization of organelles is closely linked to their biological functions, yet our understanding of higher order intracellular organization is incomplete. Here, we sought to classify breast cancer cell lines based on the spatial context of organelles within cells, specifically their subcellular location and topological inter-organelle relationships. We have introduced a novel approach that quantifies, for the first time, the topological features of subcellular organelles, removing the bias of visual interpretation, to classify different breast cancer cell lines. This method was tested on three different organelle datasets: mitochondria (Mito), early endosomes (EEC) and endosome recycling compartment (ERC) in a panel of human breast cancer cells, and non-cancerous mammary epithelial cells. A morphometric evaluation of EEC, ERC and Mito resulted in 34 topology and morphology parameters. Application of Random Forest machine learning (ML) to 18 of these 34 parameters generated the highest accuracy in breast cancer cell classification. We systematically evaluated how different parameter combinations affected the machine learning-based cancer cell classification and discovered that topology parameters were crucial to achieve a classification accuracy over 95% of human breast cancer cell lines of differing subtype and aggressiveness. These findings lay the groundwork for using quantitative topological organelle features as an effective method to analyze and classify breast cancer cell phenotypes.
Organelle compartments as the cellular proteome are highly regulated in a spatiotemporally manner. However, advanced understanding on the cellular distribution of a network of organelle compartments, i.e. organelle topology, is lacking. Here, we present OTCCP (Organelle Topology-based Cell Classification Pipeline), using ML-based method for the classification of breast cancer cell lines. To overcome the above limitations and lay the foundation for future cell recognition and diagnostics based on single cell analysis, OTCCP encompasses three major steps: (1) images were obtained by Airyscan high resolution microscopy at subcellular level and 3D rendered; (2) topology features for hundreds of organelle objects per cell were calculated; and (3) cell classification based on topology features was carried using ML algorithm. OTCCP was applied to three common organelles EEC using anti-EEA1 immunostaining, ERC using fluorescently labeled transferrin and Mito using anti-TOM20 immunostaining among six breast cancer cell lines MCF10A, AU565, MDA-MB-231 (MDA231), MDA-MB-436 (MDA436), MDA-MB-468 (MDA468), AU565 and T47D. These three different organelle topology datasets were tested using the same algorithm pipeline, outperforming topology and morphology-based methods. Based on the spatial distribution of organelles, i.e. distance from neighbor organelles (topology), a ML-based classification accuracy over 95% was achieved to discriminate between several human breast cancer cell lines of differing subtype and aggressiveness.
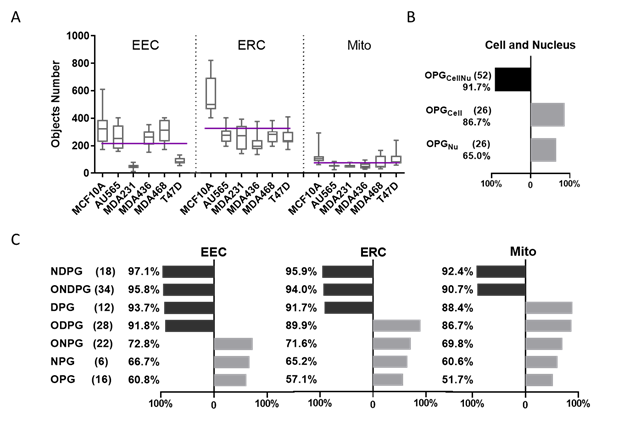
Here, we have defined organelle networks by their organelle topology, i.e. the connectivity and spatial distribution as determined using the distance between each organelle object and all of its neighbors. OTCCP obtained the highest classification accuracy for organelle datasets using NDPG: 92.4% (Mito), 95.9% (ERC), and 97.1% (EEC). Using all 34 parameters (ONDPG), OTCCP displayed reduced classification accuracy at 90.7% (Mito), 94.0% (ERC), and 95.8% (EEC). OTCCP obtained the lowest classification accuracy 51.7% (Mito), 57.1% (ERC), and 60.8% (EEC) using OPG. From highest to lowest, classification accuracy ranking is: NDPG > ONDPG > DPG > ODPG > ONPG > NPG > OPG. Among the three organelles, when 7 different parameter groups were used in the classification tasks, EEC datasets always showed the highest classification accuracy comparing to the other two organelle datasets. Mito datasets always showed the lowest accuracy.
We confirmed the importance of cellular and nuclear morphology in breast cancer cells heterogeneity in using OTCCP. Furthermore, we demonstrated that organelle topology is more important than organelle and cell morphology in the classification of breast cancer cells. Importantly, three different organelle network datasets show consistent results in their ability to classify different breast cancer lines with high accuracy. These results suggest that EEC due to their puncta nature are ideal for cell classification. We also found that positioning of endosomes and distance between endosome objects across the cell is established for each cell in a regulated manner. Thus, reinforcing our principle that organelle’s spatial distribution (topology) plays a key role in breast cancer cell classification. This notion was also confirmed by the importance index ranking, in which topology-based parameters are dominant in the top 10 parameters. These findings lay the groundwork for using organelle profiling as a potentially fast and efficient method for phenotyping breast cancer function as well as identifying other cell types and conditions.
Figure 1. Visual descriptions of different groups and their specific parameters. The pink dot represents the origin of the cell, which is the geometric center of nucleus, the blue dots represent the geometric center of each 3D rendered object. Object-based group (OPG), nucleus-related group (NPG), and distance-related group (DPG) includes 16, 6 and 12 different parameters respectively.
Figure 2. Machine learning classification comparison using cellular and subcellular morphological parameters in breast cancer cells.
A. Each cell line’s 3D rendered organelle object number. The purple line shows the mean of each organelle’s object number through 6 cell lines per cell, EEC mean = 215, EEC mean = 325, Mito mean = 76.
B. Classification accuracy by Random Forest algorithm using different parameter groups at cellular level (The black bar shows the accuracy greater than 90%, the gray bar shows the accuracy smaller than 90%, same as Fig. 2C).
C. Classification accuracy by the Random Forest algorithm using different parameter groups at subcellular level in 3 organelle datasets including EEC, ERC and Mito.
- References
Zardavas, D., Irrthum, A., Swanton, C. & Piccart, M. Clinical management of breast cancer heterogeneity. Nature Reviews Clinical Oncology (2015) doi:10.1038/nrclinonc.2015.73.
Thul, P. J. et al. A subcellular map of the human proteome. Science (80-. ). (2017) doi:10.1126/science.aal3321.
Chang, A. Y. & Marshall, W. F. Organelles – understanding noise and heterogeneity in cell biology at an intermediate scale. J. Cell Sci. 130, 819–826 (2017).
Warren, A. et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat. Commun. 12, 1–12 (2021).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature (2017) doi:10.1038/nature22369.
Collinet, C. et al. Systems survey of endocytosis by multiparametric image analysis. Nature 464, 243–249 (2010).
Zahedi, A. et al. Deep Analysis of Mitochondria and Cell Health Using Machine Learning. Sci. Rep. 8, 1–15 (2018).