Near infra-red branding (NIRB) in tissue: an efficient guide between different imaging modalities in 3D
- Abstract number
- 52
- Presentation Form
- Poster
- DOI
- 10.22443/rms.elmi2021.52
- Corresponding Email
- [email protected]
- Session
- Poster Session 1
- Authors
- Anna Kremer (1, 2, 3), Evelien Van Hamme (1, 2, 3), Peter Borghgraef (1, 2, 3), Christopher Guerin (1), Saskia Lippens (1, 2, 3)
- Affiliations
-
1. VIB BioImaging Core
2. Center for Inflammation Research, VIB
3. Department for Biomedical Molecular Biology, Ghent University
- Keywords
- NIRB
- CLEM
- 3D
- tissue
- Abstract text
Correlative Light and Electron Microscopy (CLEM) aims to image the exact same region in LM and EM with the goal to correlate ultrastructural detail to a fluorescent signal. With the development of volume-EM techniques like SBF-SEM and FIB-SEM, correlation in 3D has become more feasible. Volume electron microscopy allows for the automated acquisition of serial section imaging data that can be reconstructed in 3D to provide a detailed, comprehensive and accurate view of cellular ultrastructure. In addition, combining volume EM with super resolution techniques, decreases the resolution gap between LM and EM, making retracing of the ROI and eventual overlays more straightforward. Still, due to the difference in sample preparation between LM and EM, clear points of recognition (landmarks/fiducials) are needed to be able to retrace the region imaged by light microscopy in the electron microscope. Finally, after acquiring both LM and EM datasets, a second type of landmarks is needed to overlay both datasets correctly and make the final correlation.
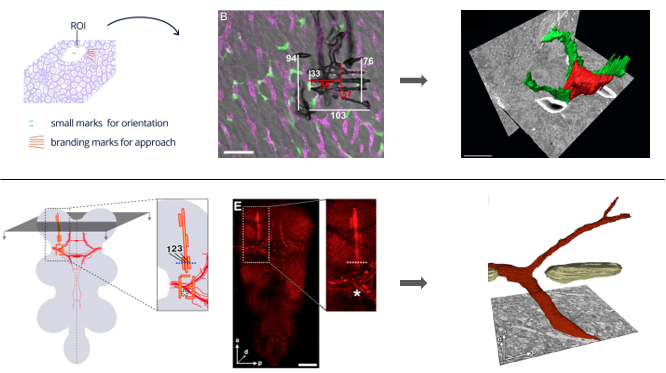
Here, we present our strategy for 3D CLEM on tissue samples imaged by super resolution confocal microscopy in combination with SBFSEM and/or FIBSEM. In these workflows, we have made use of two types of landmarks: first, we need landmarks to find back the cell or region imaged in LM in the electron microscope and second landmarks or fiducials independent of the cell or structure of interest are needed to make overlay images of LM and EM data. Near infra-red branding with a 2-photon laser in specific geometric patterns was applied to guide us in the electron microscope to the ROI in mouse liver and Drosophila brain [1, 2, 4]. Detailed knowledge on the composition and measurements of the branded pattern allows to safely approach the region of interest in the electron microscope without the risk of removing parts of the sample that contain the ROI to be imaged. Depending of the structure of the tissue, patterns can be custom-designed to rule out possible confusion in the EM images between the landmarks and tissue-specific structures, like for example blood vessels. The figure below demonstrates how different patterns were used in mouse liver and Drosophila brain to unambiguously identify the ROI. For correctly overlaying LM and EM data extra fiducials are needed. These are largely tissue dependent as well as dependent on the precision needed for the overlay (cell vs organelle). For overlaying LM and EM 3D image stacks in mouse brain we used fluorescent labeling of the blood vessels. This is illustrated in purple in the top panel of the figure below.
In this study we demonstrate how NIRB can be applied and even required to follow the ROI during the elaborate steps going from LM over EM sample preparation to 3D EM image acquisition. This approach allowed us to make 3D-CLEM overlays and correlate the fluorescent signal to the ultrastructural detail provided by the electron microscope [2, 3, 4].
Figure: NIRB in tissue. Top panel: Pattern and measurements of landmarks applied in mouse liver to guide the electron beam to the ROI defined in LM, leading to a successful overlay of LM and EM data. Bottom panel: NIRB patterns in Drosophila brain mark the dendrite branching point of interest.
- References
[1] Bishop, D., Niki´c, I., Brinkoetter, M. et al. (2011) Near-infrared branding efficiently correlates light and electron microscopy. Nat. Methods 8(7), 568–570.
[2] Bonnardel, J., T’jonck, W., Gaublomme, D. et al. (2019) Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51(4), 638–654 e9.
[3] Kremer, A., Van Hamme, E., Bonnardel, J. et al. (2020). A workflow for 3D-CLEM investigating liver tissue. J. Microsc. doi: 10.1111/jmi.12967.
[4] Urwyler, O., Izadifar, A., Dascenco, D. et al. (2015) Investigating CNS synaptogenesis at single-synapse resolution by combining reverse genetics with correlative light and electron microscopy. Development 142(2), 394–405