Medical image registration and pixel classification for the study of protein co-localization and morphology heterogeneity in cancer biopsies
- Abstract number
- 17
- Presentation Form
- Poster
- Corresponding Email
- [email protected]
- Session
- Poster Session 2
- Authors
- Laura Nicolás-Sáenz (1, 3), Nerea Carvajal (2), Javier Pascau (1, 3), Federico Rojo (2), Arrate Muñoz-Barrutia (1, 3)
- Affiliations
-
1. Departamento de Bioingenieria e Ingenieria Aeroespacial, Universidad Carlos III de Madrid, 28911 Leganes, Spain
2. Department of Pathology, CIBERONC, UAM, Fundación Jiménez Díaz University Hospital Health Research Institute, Madrid, Spain
3. Instituto de Investigación Sanitaria Gregorio Marañon, 28007 Madrid, Spain
- Keywords
computational pathology; image registration; antigen segmentation; cancer; smart pathology; automatic; non-supervised; breast cancer; Quantitative imaging; machine learning; workflow; smart microscopy; software; analysis
- Abstract text
We present a novel method to assess the variations in morphology protein expression and spatial heterogeneity of tumor biopsies with application in computational pathology. Different antigen stains in each tissue section are combined, applying complex image registration followed by a final pixel classification step, obtaining the exact location of the proteins of interest. Accurate image registration, necessary for the correct assessment of the antigen patterns, is a difficult process in histopathological images for three main reasons: the high number of artifacts due to the complex biopsy preparation, the large image size, and the complexity of the local morphology. Our previously published method [1] manages to accurately register the tissue cuts and segment the positive antigen areas. In this work, we have further proved the robustness of our solution on a new dataset of breast cancer biopsies, adding a quality measure based on the tissue artifacts, including automatic piece-based registration, and introducing segmentation for all kinds of stains.
Method:
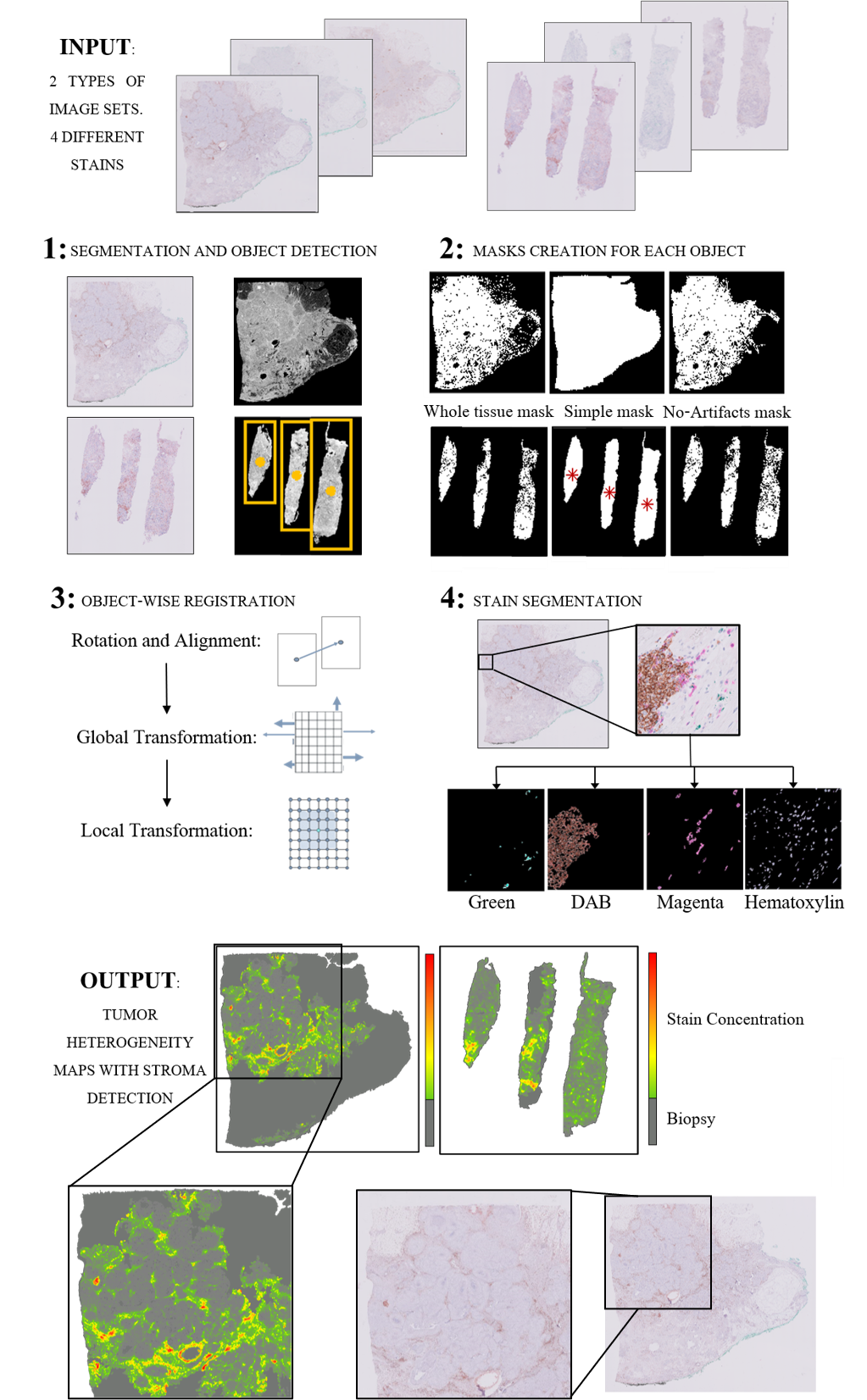
Our method consists of 4 steps: First, a robust segmentation with object detection, which is then followed by an object-wise registration. The registered images are then subjected to pixel classification for stain segmentation. Finally, the masks for the different stains are used to study morphology variations and protein-colocalization.
The segmentation of the tissue from the background is obtained analyzing L intensity level from the LAB color space histogram. In tumor biopsies, it is common to have two types of tissue sources: surgical biopsies - which include generally one big block of tissue, and needle biopsies, consisting of several slim pieces of tissue. For this reason, our method detects the number of objects in the biopsy in the segmentation mask and decides whether to proceed with the registration as a single block or for each slim piece individually.
Once the image is segmented, 3 different binary masks are created for each detected object: A simple one that takes the object as a whole mass without taking into account holes and artifacts (Simple mask), a complex one that involves the careful segmentation of all the minute structures (Whole-tissue mask), and a third one that detects artifacts, holes and fat-predominant areas and will be used for registration quality control (No-artifacts mask).
The registration algorithm calculates a transformation T consisting of a robust pre-alignment and a global and a local transformation. The combined transformation T can be expressed as T=TAlign*TGlobal*TLocal. The alignment and global transformation describe the overall motion of the biopsy and are calculated on the first segmentation mask (Simple mask). The local motion involves the alignment of the internal structure of the biopsy and is applied to the second type of mask (Whole-tissue mask). Further refinement is then calculated using the greyscale image of the segmented tissue. The third mask (No-artifacts mask) is then used to assess the areas where the registration may be faulty (folds, rips, holes, and predominately-fatty areas), obtaining a pixel-wise quality control of the registration step.
The pixel classification for stain segmentation algorithm is based on the geometry of the different color spaces’ histograms of the images. This algorithm is applied to the already registered images after applying the third mask (No-artifacts mask), thus, obtaining the segmented stains only in those areas that will be useful for the creation of the heterogeneity map. The registered stain masks are used together to form the heterogeneity map.
Results:
The final Heterogeneity Map is created from the stains segmented on the registered tissue cuts. Each transformed section is subjected to our pixel classification for stain segmentation algorithm, which results in a binary mask of the areas positive for the antigen. These masks are overlaid over the fixed image of the patient, obtaining a heterogeneity map that shows protein expression throughout the biopsy. The maps show the antigen distribution in each tissue cut with one simple and easily-to-interpret image. The hot spots of these maps would coincide with the aberrant morphology characteristics of stroma and tumor development as annotated by a pathologist (as seen in Fig. 1).
Conclusion
The proposed pipeline is robust and, most importantly, automatic and non-supervised. Compared to other segmentation and registration algorithms, our solution yields similar registration results and improved segmentation results without requiring manual annotations nor training, as shown in [1]. The obtained tumor maps provide a helpful visual representation of the intra-tumor environment and heterogeneity of the tissue. By showing the exact location of the stains within the tissue and how different dyes may correlate, this tool presents a new approach to study the tumor microenvironment. Moreover, these maps would aid in assessing tumor biopsies by automatically detecting the stroma of the tumor, identifying interesting areas and slices, or even as a tool to speed up biopsy analysis for cancer diagnosis. This could reduce diagnostic times with more accurate decisions; the use of these maps would be highly beneficial for speeding up and relieving the clogged system by making the whole process more efficient. These maps can also serve as automatic, non-supervised computer-generated input for training deep convolutional neural networks (DCNNs). Nowadays, these methods rely on manual annotations for training, which means they are dependent on the availability of pathologists. With our maps, tumoral areas and normal tissue can instead be automatically segmented and DCNNs can be trained to detect the corresponding structural information in clinical settings automatically.
Fig. 1 Process for the creation of the tumor heterogeneity maps for the two different types of images included in the breast cancer dataset: surgical biopsies and needle biopsies. In the output of the algorithm, it can be appreciated how the hotspots of the biopsies’ staining correspond to clinician-annotated areas of morphology aberrations, specifically the stroma surrounding the cluster morphology of the intratumoral space.
- References
[1] Nicolás-Sáenz, L.; Guerrero-Aspizua, S.; Pascau, J.; Muñoz-Barrutia, A. Nonlinear Image Registration and Pixel Classification Pipeline for the Study of Tumor Heterogeneity Maps. Entropy 2020, 22, 946.
https://doi.org/10.3390/e22090946