Generalized Statistical Object Distance Analysis (GSODA) For Object Based Colocalization In Quantitative Microscopy
- Abstract number
- 59
- Presentation Form
- Poster
- Corresponding Email
- [email protected]
- Session
- Poster Session 2
- Authors
- Suvadip Mukherjee (1, 3), Thibault Lagache (1, 3), Lydia Danglot (2, 4), Jean-Christophe Olivo-Marin (1, 3)
- Affiliations
-
1. Institut Pasteur
2. Paris University
3. CNRS UMR 3691
4. Inserm U1266
- Keywords
[1] Lagache, T., Grassart, A., Dallongeville, S., Faklaris, O., Sauvonnet, N., Dufour, A., Danglot, L., Olivo-Marin, J.-C., "Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics," Nature Communications, 9,1, pp. 698-711. doi: 10.1038/s41467-018-03053-x.
[2] Mukherjee, S., Gonzalez-Gomez, C., Danglot, L., Lagache, T., & Olivo-Marin, J. C. (2020). Generalizing the Statistical Analysis of Objects’ Spatial Coupling in Bioimaging. IEEE Signal Processing Letters, 27, 1085-1089.
- Abstract text
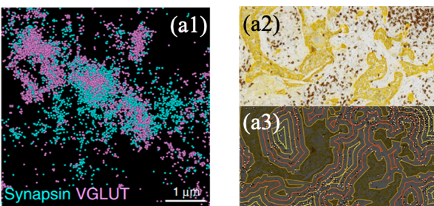
We propose a novel method to measure statistical colocalization in the paradigm of level set analysis. This solution builds on SODA [1], which is a tool to study colocalization between two (or more) spatial point processes (see Fig. 1(a1)). The proposed technique, namely Generalized SODA (GSODA) extends the applicability of its predecessor to accommodate more generic region based colocalization studies. An example application in digital pathology is illustrated in Fig. 1(a2)-(a3). GSODA provides a fast, generic, and robust model to study colocalization for a variety of problems commonly encountered in fluorescent microscopy, super resolution imaging, and histopathology. In this context, we leverage the flexibility of level sets [2] to implicitly embed closed region boundaries, and develop a mathematical model to analytically estimate the model parameters to describe the colocalization properties of the underlying random process. GSODA preserves the statistical characteristics of SODA, but extends its use to a wider gamut of colocalization studies. Furthermore, by restricting the level set function to the region of interest by using suitable boundary conditions, GSODA eliminates the need to explicitly correct for edge-artifacts, which could be challenging to compute for complex shapes. For applications involving significant spatial clustering of points, the GSODA model could be trivially extended to eliminate the need for non-trivial data unmixing, which makes the proposed solution both robust and computationally efficient. Finally, due to the recent advances in real-time polygon based digital morphology, GSODA allows computationally efficient solution for processing big data in bio-image informatics applications. Preliminary experiments suggest robustness and computational efficiency of GSODA in colocalization studies in biology.
Fig. 1: Two potential applications of object based colocalization is presented here. The first example (a1) involves estimating protein colocalization in super-resolution microscopy. In (a2), an example is taken from histopathology where the objective is to study the spatial colocalization of the immune cells (points) to the tumor regions (a3).