FRET-FLIM based dynamic screening of signal transduction pathways: a proof-of-concept study
- Abstract number
- 65
- Presentation Form
- Poster
- Corresponding Email
- [email protected]
- Session
- Poster Session 1
- Authors
- Olga Kukk (1), Rolf Harkes (1), Sravasti Mukherjee (1), Jeffrey Klarenbeek (1), Bram van den Broek (1), Kees Jalink (1)
- Affiliations
-
1. Netherlands Cancer Institute
- Keywords
cell signaling, FLIM, FRET, cAMP, β-adrenergic receptor, PDEs, deep learning, dynamic screening
- Abstract text
In this study, we present a FRET-FLIM based genetic screening platform for monitoring the dynamics of cellular cAMP (cyclic adenosine monophosphate) breakdown in real time.
Genetic screens are instrumental in identifying different gene products that contribute to a biological response. However, conventional screening methods are mostly restricted to ‘static’ end-point readouts that rely solely on the magnitude of the response. Since the progression of the response over time is also crucial in determining the final biological outcome, it is imperative that we study ‘dynamic’ events in genetic screens. Studying such dynamic responses in real-time is enabled by Forster Resonance Energy Transfer (FRET) sensors with Fluorescence Lifetime Imaging (FLIM) being the most quantitative method to readout FRET. However, until recently, FRET detection lacked the robustness and speed necessary to extract quantitative data from single cells. Thus, with highly sensitive and photon-efficient FLIM instrumentation and a dedicated FRET-FLIM biosensor, we set out to demonstrate a dynamic genetic screening platform and automated analysis pipeline for monitoring the activity of proteins involved in cellular signal transduction, taking the cAMP pathway as an example.
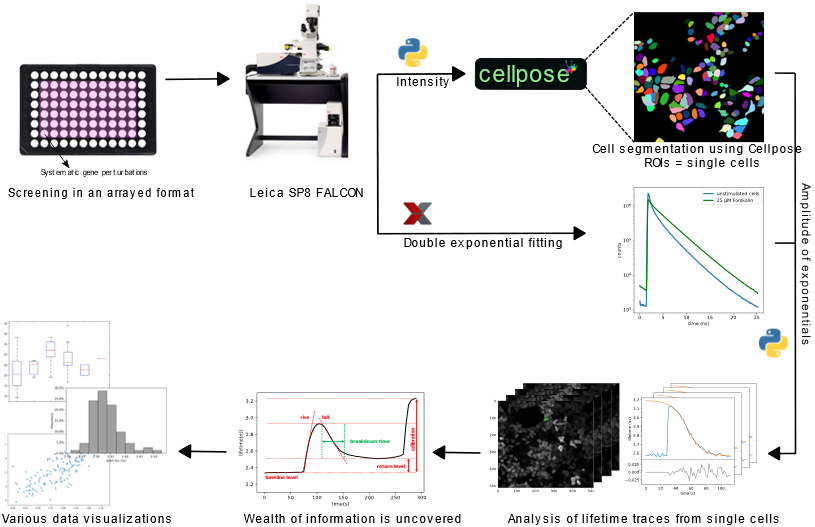
Using HeLa cells stably expressing our Epac based FRET sensor1, we investigated the effects of siRNA-mediated individual knockdown of a set of 22 different phosphodiesterases (PDEs) on cAMP breakdown kinetics in a multi-well format. For analyzing the screens, an automated analysis workflow was developed using custom Python scripts. Fluorescence lifetimes were measured by time correlated single photon counting using the Leica FALCON system2. Cell segmentation was performed by a deep-learning algorithm, Cellpose3. The lifetime traces for all individual live cells were then fitted to a logistic function to obtain the cAMP breakdown rate corresponding to the cellular PDE activity (Figure 1).
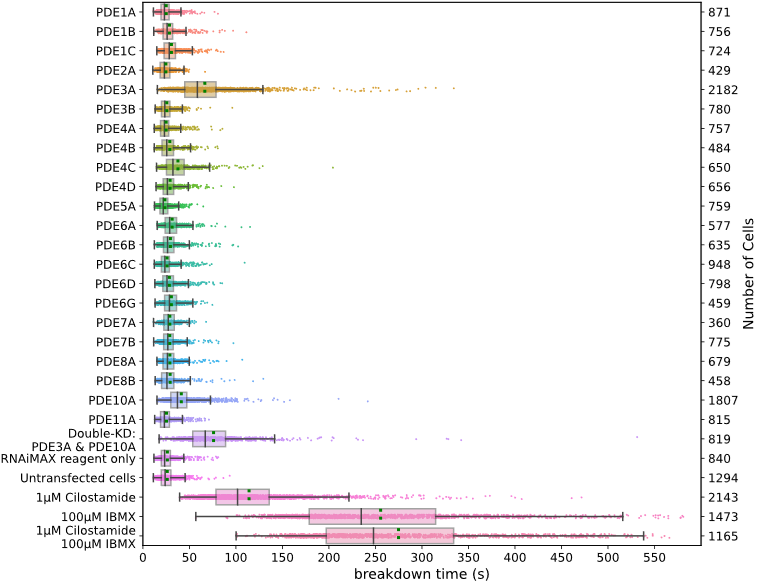
Our FRET-FLIM screening pipeline produces a wealth of robust and reproducible data on cAMP metabolism. We identify PDE3A and PDE10A as the dominant PDEs that determine cAMP breakdown rates following cAMP production, whereas their knockdown has little or no effect on basal cAMP levels in HeLa cells (Figure 2). Additionally, this study demonstrates that FLIM recording has now become sufficiently fast and sensitive for single cell dynamic screening experiments, thus making FLIM a very attractive choice for FRET-based signaling studies.
In conclusion, we present a robust FLIM-enabled screening platform that provides detailed kinetic analysis of cellular signals in individual cells. This platform will also pave the way for performing genome-wide screens focused on dynamics of signal transduction under physiologically relevant conditions.
Figure 1: Schematic overview of the FLIM screen for monitoring dynamic changes in cAMP
Figure 2: Breakdown of cAMP for different knockdowns of PDEs upon brief stimulation of the β-adrenergic pathway. Datapoints are fitted decay times of single cells. For each condition, the experiment was performed in duplicates, with cells grown, transfected, and assayed in two independent wells. Indicated are median value (vertical black line), mean value (green dotted line); boxes encompass middle 50% of values and whiskers represent 1.5 times the interquartile range.
- References
1. Klarenbeek, J.B., Goedhart, J., Hink, M.A., Gadella, T.W. and Jalink, K., 2011. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PloS one, 6(4), p.e19170.
2. Alvarez, L.A., Widzgowski, B., Ossato, G., van den Broek, B., Jalink, K., Kuschel, L., Roberti, M.J. and Hecht, F., 2019. SP8 FALCON: A novel concept in fluorescence lifetime imaging enabling video-rate confocal FLIM. Nat. Methods, 16, pp.1069-1071.
3. Stringer, C., Wang, T., Michaelos, M. and Pachitariu, M., 2021. Cellpose: a generalist algorithm for cellular segmentation. Nature Methods, 18(1), pp.100-106.