Dynamics of chromatin proteins RSF1, CENPS and CENPX at laser induced DNA double strand breaks
- Abstract number
- 70
- Presentation Form
- Poster
- DOI
- 10.22443/rms.elmi2021.70
- Corresponding Email
- [email protected]
- Session
- Poster Session 1
- Authors
- Pritishkumar Tidke (2, 1), Andrew Flaus (1), Helen Dodson (2)
- Affiliations
-
1. Centre for Chromosome biology, Biomedical Sciences Building, National university of Ireland, Galway, Ireland
2. Department of Anatomy, National university of Ireland, Galway, Ireland
- Keywords
RSF1, CENPS, CENPX, DNA damage response, Live cell quantitative microscopy, kinetics study, laser microirradiation, cell cycle phases.
- Abstract text
Chromatin has a major influence on the DNA damage response (DDR). Several chromatin-related factors contribute to specialized DNA packaging during the DDR including the CENPS and CENPX histone fold proteins (also known as MHF1/2) and the chromatin remodelling factor RSF1. We hypothesised that CENPS/X could contribute to an intermediate and more accessible state of chromatin packaging that facilitates the resection and repair process. To establish the dynamic exchanges of these chromatin components during the DDR we have defined a timeline for recruitment and removal of MRE11, RSF1, CENPS, and CENPX at induced DNA double strand breaks (DSBs).
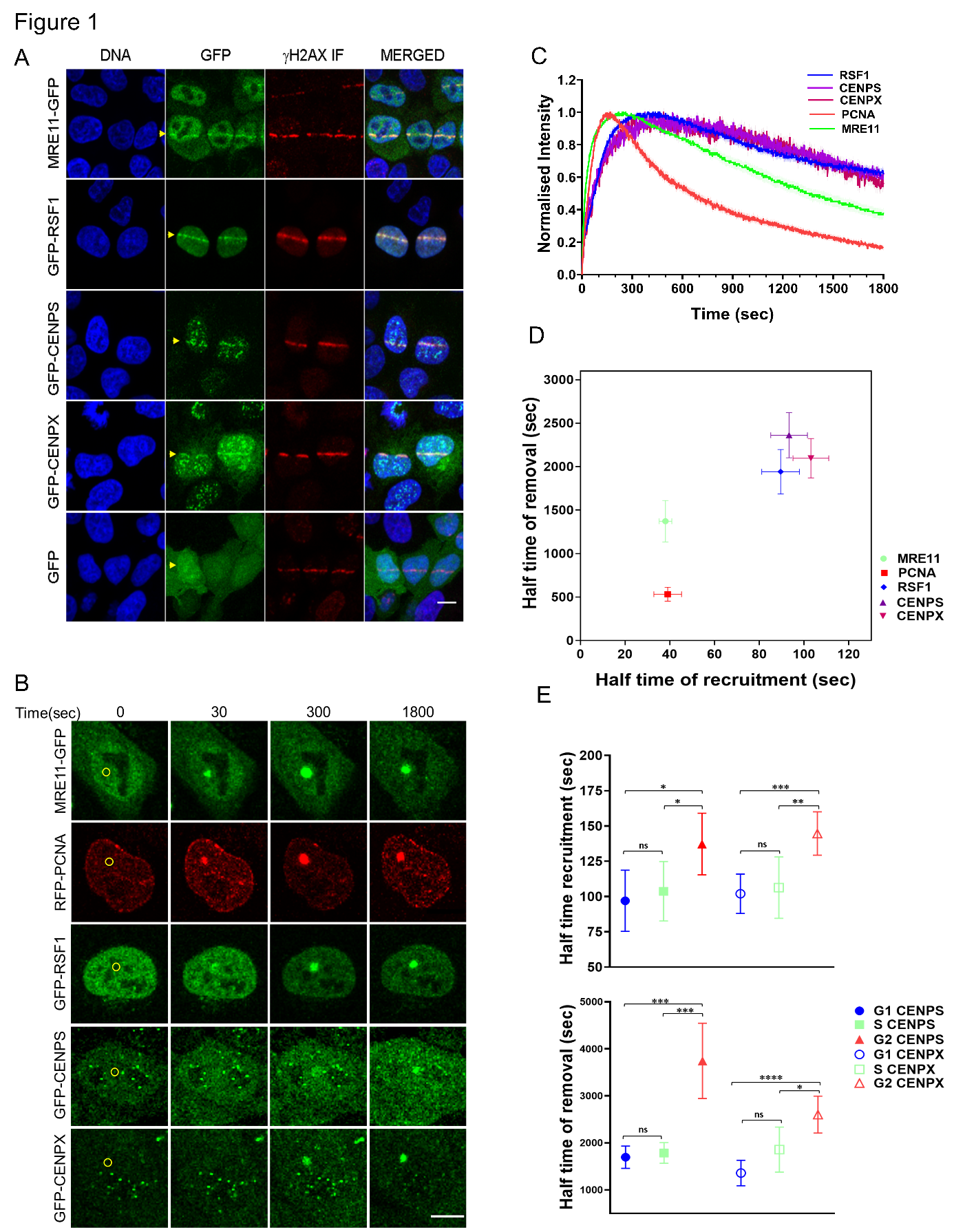
Stable expression of GFP-tagged CENPS, CENPX and RSF1 was established in HeLa Kyoto (HK) cell lines by transfection and selection in order to observe the DDR in real time by laser microirradiation followed by live confocal imaging. We explored mild conditions to induce DSBs in HK cells using a chemical sensitiser and 405 nm laser microirradiation and found that 2 µM Hoechst-33342 treatment followed by 0.5 mW laser power created single foci of recruitment at the sites of microirradiation while other combinations had insufficient fluorescence intensity or resulted in excessive damage with non-specific foci outside the region of interest. The induction of DSBs at sites of microirradiation was validated by co-localisation of gH2AX immunostaining (Figure 1A).
Fitting a consecutive chain reaction model to the observed time course of fluorescence at microirradiated foci then enabled quantitative estimation of the half time of recruitment (K1HT) and removal (K2HT) for GFP-tagged MRE11, RSF1, CENPS and CENPX using at least 20 measurements from five biological replicates. MRE11 recruitment and removal (K1HT 38 ± 6 sec; K2HT 1370 ± 470 s) is consistent with reported values for the MRN complex and precedes RSF1, CENPS and CENPX (K1HT 89 ± 16, 93 ± 16, 103 ± 16 s; K2HT 1940 ± 500, 2360 ± 510 and 2100 ± 440 s respectively) (Figure 1B-D). The three chromatin components are recruited and removed at a similar time to each other within the resolution of the measurements. This locates the chromatin-related components at DSBs at a time subsequent to non-homologous end joining (NHEJ) components observed by others using the same method, and with recruitment and removal times that immediately precede the homologous recombination (HR) machinery.
A caveat to these measurements is that the cell cycle phase is unknown and likely to be dominated by G1 cells. To study the cell cycle phase specific association of CENPS and CENPX at DSBs, we prepared dual tagged HK cell lines stably expressing RFP-PCNA and either GFP-CENPS or GFP-CENPX. These cells were synchronised by double thymidine block and microirradiated at optimal times for enrichment in S, G2 or G1 phases. We observed that CENPS and CENPX are recruited to DSBs in all phases of the cell cycle, but that the half time of their recruitment and removal in G2 phase (K1HT 137 ± 24 and 144 ± 15s; K2HT 3710 ± 870 and 2600 ± 390 s respectively) is somewhat delayed compared to G1 and S phase (K1HT 106 ± 23 and 108 ± 23 s; K2HT 1650 ± 290 and 1690 ± 190 s respectively) (Figure 1 E).
We also estimated the abundance of CENPS and CENPX recruited to the focus by normalising to the total available fluorescent signal in each cell before microirradiation. This revealed that G2 recruitment is three times higher than in G1, which explains the apparent delay in G2 since the time to achieve half maximal abundance would be longer at constant recruitment and removal rates.
Previous work has shown that CENPS and CENPX bind simultaneously to two DNA duplexes as CENPS/X heterotetramers, and that they are recruited to DSBs by the chromatin remodelling factor RSF1 for efficient DSB repair, possibly linked to the association of CENPS/X with FANCM. Our observations demonstrate that CENPS/X recruitment occurs between the regimes of fast NHEJ and slower HR, at a time consistent with published estimates for ATM activation that initiates resection. The release of CENPS/X is at a similar time to the recruitment of RAD51. This place the histone fold containing DNA binding CENPS/X in the vicinity of DSBs at the time when their nucleosomes are being actively remodelled to enable resection, and increased abundance of CENPS/X at DSBs in G2 correlates with extended resection for HR. Our work provides an in vivo basis for investigating the detailed mechanistic and structural contribution of CENPS/X in the DDR.
Figure 1. Live cell quantitative microscopy of chromatin components at DSBs. A. HeLa Kyoto (HK) cells expressing GFP-fused proteins were fixed and stained with anti-gH2AX 600 s after stripe microirradiation (yellow arrowhead). Scale bar 10 μm. B. HK cells were imaged for up to 1800 seconds after spot microirradiation (yellow circle at 0 s). Scale bar 10 μm. C. Normalised fluorescence intensity at 2 s intervals at a defined ROI surrounding the microirradiation site for MRE11-GFP, RFP1-PCNA, GFP-RSF1, GFP-CENPS and GFP-CENPX averaged over 21 cells from at least 5 biological replicates. D. Half time of recruitment and removal. Error bars indicate 95% confidence interval. E. Half time of recruitment and removal of GFP-CENPS and GFP-CENPX at the microirradiation site in G1, S and G2 phases. Error bars indicate 95% confidence interval. Groups were compared by a two tailed Student’s t-test with p values indicated as * ≤0.05, ** ≤0.01, *** ≤0.001, **** ≤ 0.0001, ns = not significant.
- References