Computational tool for automated study of cell division dynamics in 3D cellular spheroids
- Abstract number
- 60
- Presentation Form
- Poster
- DOI
- 10.22443/rms.elmi2021.60
- Corresponding Email
- [email protected]
- Session
- Poster Session 2
- Authors
- Stylianos Didaskalou (2), Lito Karkaletsou (2), Christos Efstathiou (2, 1), Avgi Tsolou (2), Andreas Girod (1), Maria Koffa (2)
- Affiliations
-
1. Department of Life Sciences and Medicine, University of Luxembourg, Esch-sur-Alzette, Luxembourg
2. Department of Molecular Biology and Genetics, Democritus University of Thrace, Alexandroupolis, Greece
- Keywords
mitosis, spheroids
- Abstract text
Recent studies focus on three-dimensional cell cultures that more accurately mimic the environment of a tumor in vivo, compared to two-dimensional (monolayer) cell cultures. Light Sheet Fluorescence Microscopy (LSFM) is the gold standard for three-dimensional imaging of multicellular spheroids, providing both high spatial and temporal resolution, while maintaining low phototoxicity and photobleaching levels. Although LSFM microscopes are widely utilized for live imaging of such thick samples with subcellular resolution, the enormous amount of data generated makes manual analysis of 4D data (3D+t) challenging. To this end , we aimed to develop a pipeline in order to automatically analyze the temporal dynamics of cells divisions inside 3D multicellular spheroids.
In a multicellular spheroid, cells proliferate in close proximity, thus better mimicking cell-cell interactions, such as physical communication and signaling pathways. Moreover, the 3D geometry of spheroids restricts the flow of growth nutrients and oxygen towards the inner layers, forming proliferation gradients commonly found in tumors1. Recent studies demonstrate that nuclei inside cleared cellular spheroids are elongated, with the longest axis preferentially parallel to the spheroid’s surface2. Moreover , live imaging of spheroids shows a preference in the orientation of the mitotic spindle, with its division axis almost parallel to the surface. However, the aforementioned analysis requires manual nuclei segmentation. For this reason, our study aims to automate nuclei segmentation, cell cycle phase classification and cell tracking, in order to study cell division dynamics in the interior of 3D multicellular spheroids.
In our preliminary experiments, HeLa Kyoto cells stably expressing mCherry-H2B were grown as multicellular spheroids with an approximate diameter of 150μm. Spheroids were embedded in low melting- agarose and imaged with a light sheet fluorescence microscope (LaVision Biotech, Bielefeld / Germany). In order to segment individual nuclei of different sizes, a scale-space Laplacian of the Gaussian blob detector was used to identify seed points, followed by an immersed watershed segmentation to separate adjacent cells in the initial segmented binary image3,4.
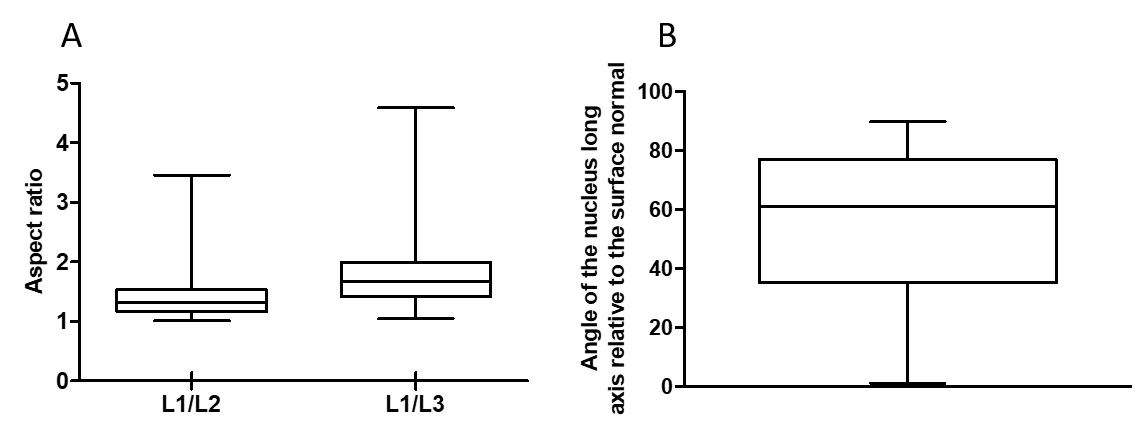
Analysis, of n=1090 nuclei derived from N =6 spheroids, showed that nuclei are elongated with an aspect ratio of (mean ± S.D) between long and short axis (Figure 1A). Additionally, using morphological operations and alpha shape to approximate spheroid surface, we were able to analyze the orientation of nuclei long axis compared to the spheroid surface. The analysis showed nuclei elongation preferentially in parallel to the surface (Figure 1B).
To further classify cells according to their cell cycle phase, a convolution neural network is used. As training a neural network requires user-annotated data, an application to simplify the process has already been developed . With this application, the user has the ability to label the already segmented nuclei according to the cell cycle phase. Finally, cells can be tracked in time, providing the opportunity to extract useful temporal information.
Advances in both fluorescence microscopy and three-dimensional cell culture methods enable us to fill the gap between 2D cell cultures and real tissues. However, the tremendous amount of generated data imposes the need for development of computational pipelines and tools for the automatic and robust analysis of high dimensionality images. This will help us study the spatiotemporal dynamics of cell division, in 3D tumor models with greater accuracy and precision.
Figure 1. A) Aspect ratio between long and middle axis (L1/L2) and long and short axis (L1/L3) of automatic segmented nuclei. B) Angle between long axis of nuclei and the normal to the surface. The normal to the surface is being computed to the surface closest point from each nucleus centroid.
- References
1. Pampaloni, F., Ansari, N. & Stelzer, E. H. K. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 352, 161–177 (2013).
2. Desmaison, A. et al. Impact of physical confinement on nuclei geometry and cell division dynamics in 3D spheroids. Sci. Rep. 8, 1–8 (2018).
3. Stegmaier, J. et al. Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks. PLoS One 9, 1–11 (2014).
4. Schmitz, A., Fischer, S. C., Mattheyer, C., Pampaloni, F. & Stelzer, E. H. K. Multiscale image analysis reveals structural heterogeneity of the cell microenvironment in homotypic spheroids. Sci. Rep. 7, 43693 (2017).