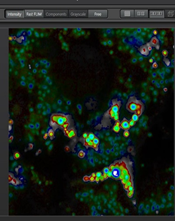
A close-up view of mitophagy using mt-keima and fluorescence lifetime microscopy (FLIM)
- Abstract number
- 4
- Presentation Form
- Oral
- DOI
- 10.22443/rms.elmi2021.4
- Corresponding Email
- [email protected]
- Session
- Live and Functional Imaging Technologies Part 1
- Authors
- Daniela Malide (2), Nuo Sun (3), Toren Finkel (1)
- Affiliations
-
1. Aging Institute, University of Pittsburgh Medical Center, Pittsburgh, PA, USA
2. NHLBI light microscopy core, NIH, Bethesda, MD, USA
3. Ohio State University Wexner Medical Center, Columbus, Ohio, USA
- Keywords
mt-keima, mitophagy, FLIM, ph-dependent fluorescence proteins
- Abstract text
Mitophagy is a cellular process that selectively removes damaged, old or dysfunctional mitochondria. Defective mitophagy is thought to contribute to normal aging and to various neurodegenerative and cardiovascular diseases. Previous methods used to detect mitophagy in vivo were cumbersome, insensitive and difficult to quantify. We created a transgenic mouse model that expresses the pH-dependent fluorescent protein mt-Keima in order to more readily assess mitophagy. Keima is a pH-sensitive, dual excitation ratiometric fluorescent protein that also exhibits resistance to lysosomal proteases [1,2]. At the physiological pH of the mitochondria (pH 8.0), the shorter-wavelength excitation predominates. Within the acidic lysosome (pH 4.5) after mitophagy, mt-Keima undergoes a gradual shift to longer-wavelength excitation. In addition to intensity imaging we describe here how to apply mt-Keima fluorescence lifetime microscopy (FLIM) to visualize mitophagy in live cells as well as various tissues including skeletal muscle, heart, liver, and kidney, obtained from mt-Keima transgenic mice. We observed that in control live cells mt- Keima fluorescence exhibits two components a short (0.4ns) lifetime corresponding to the mitophagic compartment and a longer (2.6ns) lifetime corresponding to normal mitochondria, in good correspondence to the intensity images. Interestingly, in the tissues the lifetime measurements reveal a heterogeneous mitophagic compartment containing in addition to the short (0.5ns) lifetime mt-Keima species an intermediary (1.2ns) longer lifetime component. Whether these 2 components correspond to different folding states, digestion products of the mt-Keima in the acidic environment remains to be elucidated. In conclusion FLIM provide a complementary approach to asses mitophagy in normal cells and tissues as well
as in disease situations, or altered under environmental, genetic perturbations, or in aging.
Liver-FLIM-phasor separation
- References
[1]. N. Sun; J. Yun; J. Liu, D. Malide; C. Liu; II. Rovira; KM. Holmström; MM. Fergusson; YH. Yoo; CA. Combs and T. Finkel Measuring in vivo mitophagy Mol Cell. 60(4):685-696 (2015)
[2]. N. Sun; D. Malide; J. Liu; II. Rovira; CA. Combs and T Finkel. A mouse model for measurement of mitophagy. Nature Protocols 12, 1576–1587 (2017)